11.2. Data Interpolation¶
Data interpolation algorithms are used to fill in the unknown data points between data points that are known.
11.2.1. One-Dimensional Interpolation¶
MATLAB has two functions that interpolate between one-dimensional data
samples to estimate unknown points. The difference between the two
functions relates only to the purpose for calling them. The
fillmissing function is used when values that should be in the data
are missing—often replaced with the NaN symbol. The interp1
function adds data points not contained in the data. We used
fillmissing in Dealing with Missing Data when data imported into MATLAB
had some missing values, and we will use interp1 in the examples in
this section. We call interp1 with vectors for the given \(x\)
and \(y\) data values (x1, y1), along with a longer vector
of \(x\) values (x2) where we want to find corresponding
interpolated \(y\) values. An optional fourth argument is a string
listing the interpolation algorithm.
y2 = interp1(x1, y1, x2, <algorithm>)
Several interpolation algorithms are available. Five commonly used algorithms are:
’nearest’n̄earest-neighbor interpolation’linear’linear interpolation, default’pchip’piecewise cubic Hermite interpolation’makima’modified Akima cubic Hermite interpolation’spline’cubic spline interpolation
MATLAB also has functions that directly invoke the data interpolation
algorithms rather than invoking them via fillmissing or interp1.
These functions include pchip, spline, and makima.
The default interpolation algorithm is ’linear’, which does not
usually provide smooth transitions between points but is good when the
given data changes slowly between sample points. The methods
’pchip’, ’makima’, and ’spline’ yield good results. The
’spline’ method requires the most computation because it uses a
matrix computation for each point to determine polynomial coefficients
that match the given data and maintain constant first and second order
derivatives at each data point added. However, the ’spline’ method
may overshoot data points, causing an oscillation. The results from the
’pchip’ and ’makima’ algorithms are similar. They maintain
consistent first-order but not consistent second-order derivatives at
each point. They avoid overshoots and accurately connect the flat
regions. Another advantage of the ’pchip’ and ’makima’
algorithms is that if the given data points are either monotonically
increasing or decreasing, so will the interpolated data
points [HIGHAM17]. The ’spline’ interpolation may
not always maintain data monotonicity because of oscillations. Since
’pchip’ and ’makima’ have similar results, only the ’pchip’
interpolation is used in the following comparison code and plots in
figure Fig. 11.7.
f = @(x) 1 + x.^2 - x.^3 + 20*sin(x);
x1 = (-3:1.5:3)'; % Limited data points
y1 = f(x1);
x2 = (-3:0.5:3)'; % More data points
subplot(2,2,1)
plot(x1,y1,'o',x2,interp1(x1,y1,x2,'nearest'),'+:')
title('nearest'), subplot(2,2,2)
plot(x1,y1,'o',x2,interp1(x1,y1,x2,'linear'),'+:')
title('Linear'), subplot(2,2,3)
plot(x1,y1,'o',x2,interp1(x1,y1,x2,'pchip'),'+:')
title('pchip'), subplot(2,2,4)
plot(x1,y1,'o',x2,interp1(x1,y1,x2,'spline'),'+:')
title('spline')
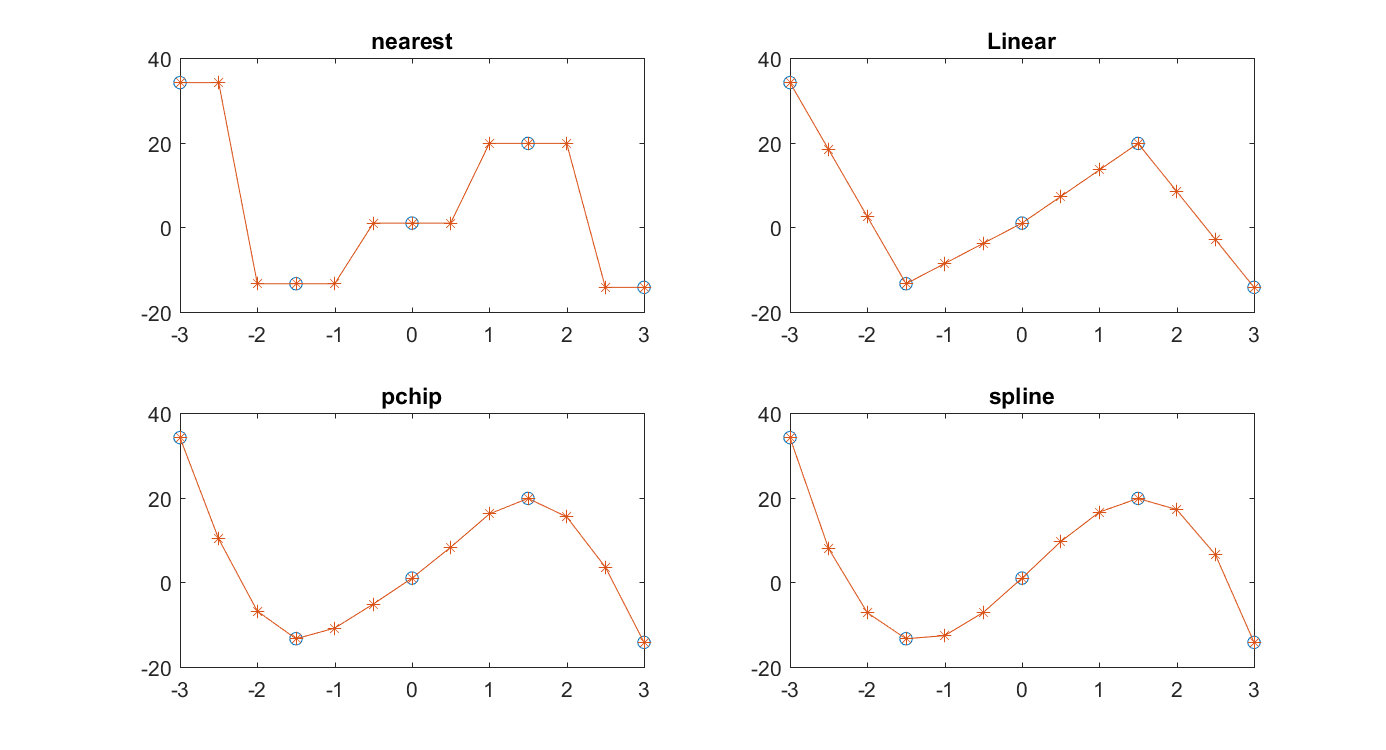
Fig. 11.7 Comparison of four data interpolation algorithms. The circles show the given data points, while the interpolated points are marked with plus signs.¶
The following example illustrates the difference between ’pchip’ and
’spline’ interpolation results. As figure Fig. 11.8
shows, the ’pchip’ result may not be as smooth, but the monotonicity
of the data is preserved.
x = -3:3;
y = [-1 -1 -1 0 1 1 1];
t = -3:.01:3;
hold on
plot(x, y, 'o')
plot(t, interp1(x, y, t, 'pchip'), 'g')
plot(t, interp1(x, y, t, 'spline'), '-.r')
legend('data', 'pchip', 'spline', 'Location', 'NorthWest')
hold off
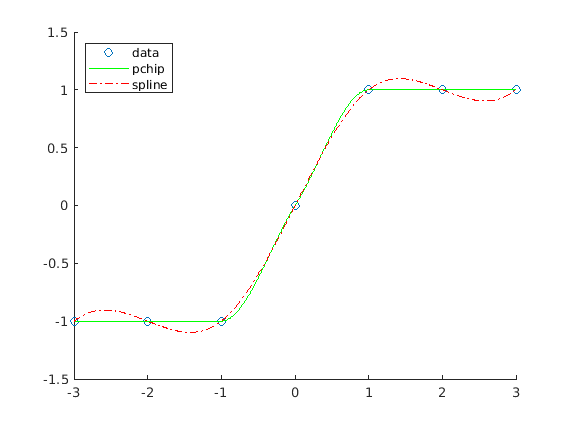
Fig. 11.8 Comparison of ’pchip’ and ’spline’ data interpolation¶
11.2.2. Two-Dimensional Interpolation¶
The two-dimensional interpolation functions griddata and interp2
follow in the same pattern as the one-dimensional functions. The inputs
are matrices of grid points, such as returned from meshgrid, which
we used in 3-D Plots to make surface plots. The distinction
between griddata and interp2 is that interp2 requires that
the grid data be monotonically increasing from meshgrid, while
griddata does not have that requirement. The available interpolation
methods are ’linear’, ’nearest’, ’cubic’, ’makima’, or
’spline’ with the default method again being ’linear’. In the
following example, we will use the default interpolation. The appearance
of the surface plot in figure Fig. 11.9 is
indistinguishable from a surface plot where all of the points come from
an equation rather than having some interpolated points.
>> [X, Y] = meshgrid(linspace(-10, 10, 20));
>> T = X.^2 + Y.^2;
>> Z = (Y-X).*exp(-0.12*T);
>> [Xi, Yi] = meshgrid(linspace(-10, 10, 40));
>> Zi = interp2(X, Y, Z, Xi, Yi);
>> surf(Xi, Yi, Zi)
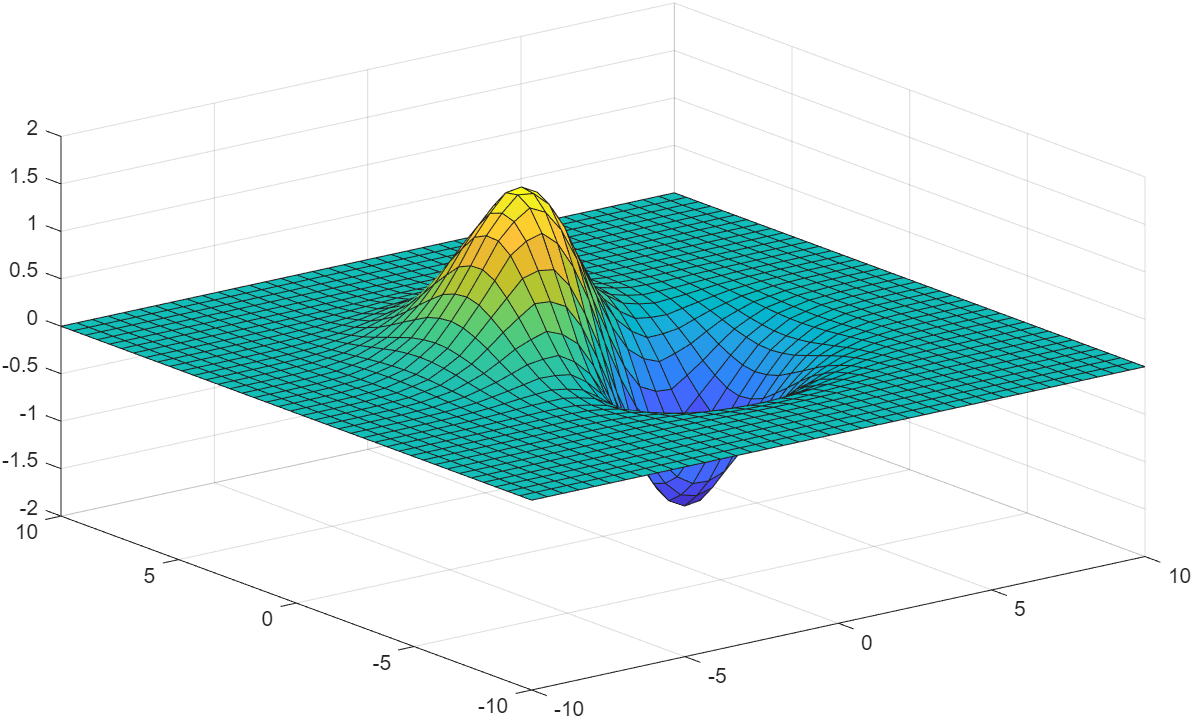
Fig. 11.9 A surface plot where half of the data points are found from linear interpolation.¶
